General information
Galaksio is a web tool that provides a simple but complete UI for using Galaxy for biologists that require bioinformatical workflows to complete their researching. Galaxy is an open source, web-based platform for data intensive biomedical research. A workflow in Galaxy is a series of tools and dataset actions that run in sequence as a batch operation.
The major steps of workflow execution using Galaksio can be summarized as follows:
- Log in the application using your Galaxy account or create a new one.
- Upload your data to Galaksio to your Galaxy history and design your dataset collections.
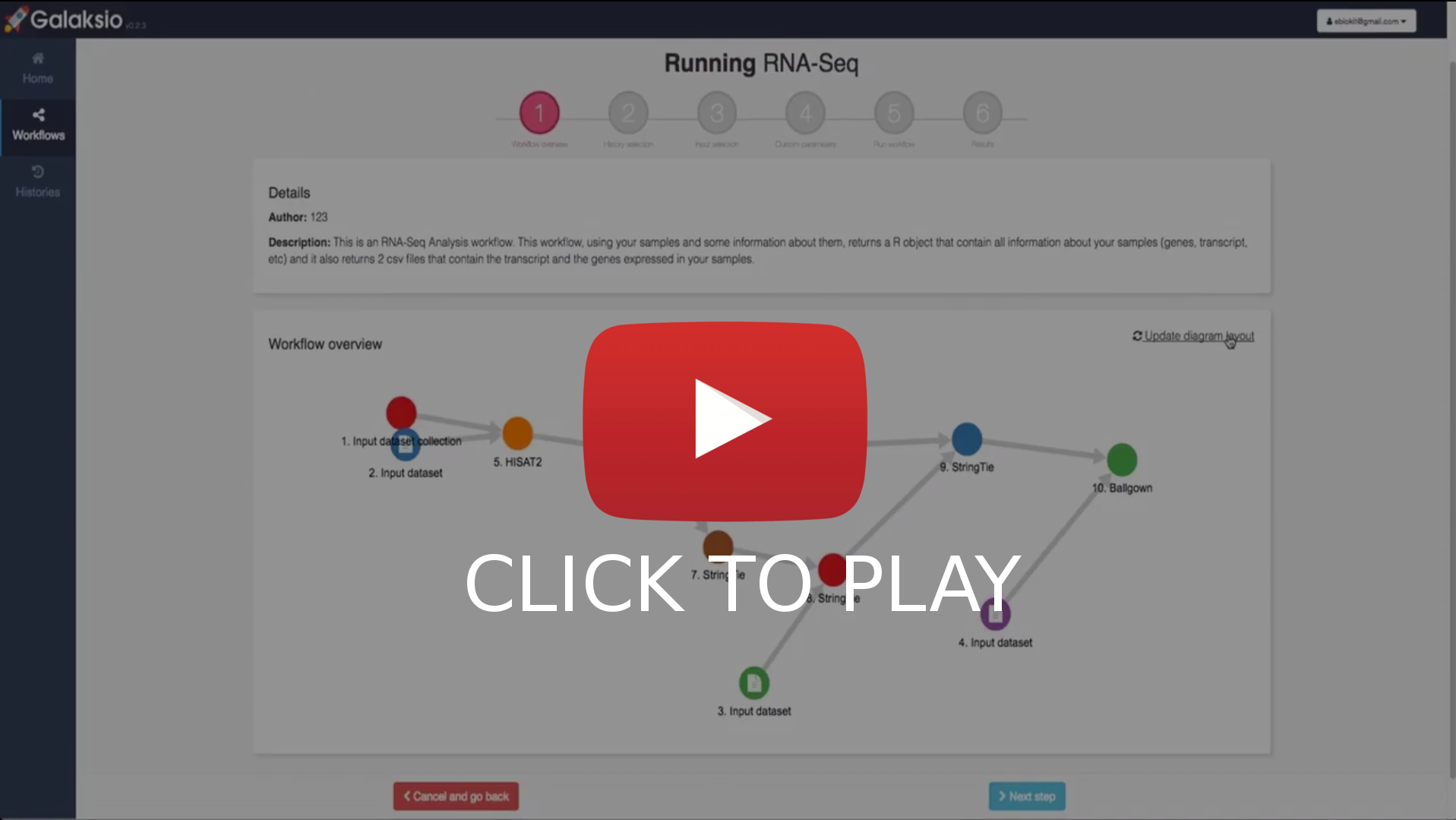
- Browse the publicly available workflows in Galaxy and import them to your collection.
- Select a workflow to run and set the input files from your history. Optionally, customize the parameters for the tools in the workflow.
- Launch the workflow execution and wait for results, or send the execution to background and continue working.
- Evaluate and download the results.
Galaksio and Galaxy
Galaksio is built to provide a more layered approach to Galaxy, providing a simplified user interface based on workflows and reducing the workload of bioinformaticians as routine tasks can be performed with minimal training.
Galaksio uses the Galaxy API to run tools and manipulate the datasets, which means that every action in Galaksio is sent and executed in a Galaxy server. This also means that user accounts in Galaksio are the same than in Galaxy. Users can use their existing accounts in Galaxy to log in Galaksio, and new accounts created in Galaksio will be also available in the Galaxy server. By default, Galaksio will send the actions to the official Galaxy server running at https://usegalaxy.org/. However, this option can be changed at any time by the administrator, using alternative Galaxy servers.
Galaksio or Galaxy? Target users for Galaksio
Galaxy is a widely supported workflow management system that provides access to bioinformatics for experimentalists with limited expertise in programming. While researchers skilled enough in bioinformatics usually prefer command line tools, less advanced users are left on their own struggling to find and combine tools using the rich but unforgiving user interface provided by Galaxy. However, even for the most simplistic use of Galaxy, insights in bioinformatics are required in order to choose an optimal combination of tools, and set their parameters correctly.
From the perspective of Galaksio, workflows implementation must be performed by skilled users (e.g. a bioinformatician) using the Galaxy tools for workflow edition. Implemented workflows are made public letting other users to reuse them. Less skilled users can access these public workflows at the "Workflows" section in Galaksio. Hence, a biologist that want to conduct his bioinformatics analysis, can search for a workflow that fullfills his requirements, and run it easily just choosing the input datasets.
What can I do with Galaksio?
|Feature | Category | Implemented | Planned| | | --- | --- | :---: | :---: | |User sign-in/out | Users | X | | |User sign-up | Users | X | | |Workflow listing | Workflows | X | | |Workflow importing | Workflows | X | | |Workflow execution | Workflows | X | | |Workflow creation | Workflows | X | NO | |Simultaneous execution of workflows | Workflows | X | | |Recovering previous executions | Workflows | X | | |Help and description for tools in workflow | Workflows | X | | |Input selection and parameter configuration | Workflows | X | | |History selection | History | X | | |History creation | History | | YES | |History deletion | History | | YES | |Dataset uploading | Dataset manipulation | X | | |Dataset downloading | Dataset manipulation | X | | |Dataset deletion | Dataset manipulation | X | | |Dataset collection creation | Dataset manipulation | X | | |Dataset collection deletion | Dataset manipulation | | YES | |Tool execution | Tools | | YES |